Spectral Analysis Exercises Solutions
Initially please make sure the course_data_dir and course_data_url variables are set:
course_data_dir = "./myEMURdata" # change to valid dir path on your system
course_data_url = "http://www.phonetik.uni-muenchen.de/~jmh/lehre/Rdf"All the Q & A’s are based on the kielread_emuDB that you can download from this location: http://www.phonetik.uni-muenchen.de/%7Ejmh/lehre/sem/ws1819/emuR/LESSON4/kielread_emuDB.zip After downloading and unpacking the emuDB place it into the course_data_dir directory.
You will also need the following R packages:
library(emuR)
library(tidyverse) # this includes dplyr and ggplot2and to load the emuDB:
kiel_db = load_emuDB(file.path(course_data_dir, "kielread_emuDB"))## INFO: Checking if cache needs update for 1 sessions and 200 bundles ...
## INFO: Performing precheck and calculating checksums (== MD5 sums) for _annot.json files ...
## INFO: Nothing to update!Q & A’s
Please place your answers in the R Markdown code blocks or write the answers next to the A:
Visualizing smoothed vowel formant trajectories
- Q: How do you get a overview/summary of the emuDB configuration?
- A:
summary(kiel_db)## Name: kielread
## UUID: 08b534b5-916a-4877-93aa-4f6b641995a1
## Directory: /tmp/WP_4.1/myEMURdata/kielread_emuDB
## Session count: 1
## Bundle count: 200
## Annotation item count: 12294
## Label count: 32978
## Link count: 11271
##
## Database configuration:
##
## SSFF track definitions:
## name columnName fileExtension
## 1 FORMANTS fm fms
## 2 dft dft dft
##
## Level definitions:
## name type nrOfAttrDefs attrDefNames
## 1 Word ITEM 2 Word; Func;
## 2 Syllable ITEM 1 Syllable;
## 3 Kanonic ITEM 2 Kanonic; SinfoKan;
## 4 Phonetic SEGMENT 4 Phonetic; Autoseg; SinfoPhon; LexAccent;
##
## Link definitions:
## type superlevelName sublevelName
## 1 ONE_TO_MANY Word Syllable
## 2 ONE_TO_MANY Syllable Kanonic
## 3 MANY_TO_MANY Kanonic Phonetic- Q: How do we add a SSFF track to the emuDB called ‘FORMANTS’ and also calculate the SSFF files using the wrassp function forest?
- A:
# ignore error if track already exists
add_ssffTrackDefinition(kiel_db,
name = "FORMANTS",
onTheFlyFunctionName = "forest")## Error in add_ssffTrackDefinition(kiel_db, name = "FORMANTS", onTheFlyFunctionName = "forest"): ssffTrackDefinitions with name FORMANTS already exists for emuDB: kielread!- Q: How do we extract the a: and i: vowels from the database?
- A:
sl = query(kiel_db, "Phonetic == a:|i:")- Q: How do we extract the formant values for the previously extracted vowels?
- A:
td = get_trackdata(kiel_db, sl, ssffTrackName = "FORMANTS")##
## INFO: parsing 139 fms segments/events
##
|
| | 0%
|
|= | 1%
|
|== | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========== | 14%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|=============================== | 45%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================= | 71%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|====================================================== | 78%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================= | 94%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%- Q: How do we apply a dct-smoothing (m=2) of the F1 and F2 tracks?
- A:
td_smooth = td %>%
group_by(sl_rowIdx) %>%
mutate(F1_smoothed = emuR::dct(T1, m = 2, fit = T), F2_smoothed = emuR::dct(T2, m = 2, fit = T))- Q: Bundles that start with K67 were produced by a male speakers and bundles that start with K68 where produced by a female speaker. How do we add a column to our trackdata called ‘sex’ that has the values ‘male’ and ‘female’ for the respective bundles?
- A:
td_smooth_gender = td_smooth %>%
mutate(sex = str_replace(bundle, "K67.*", "male")) %>%
mutate(sex = str_replace(sex, "K68.*", "female"))- Q: How do we visualize the dct-smoothed F1 and F2 tracks (one panel for each vowel category and sex)?
- A:
# plot the smoothed slices
ggplot(td_smooth_gender) +
aes(x = times_norm, y = F1_smoothed, group = sl_rowIdx) +
geom_line() +
geom_line(aes(y = F2_smoothed, col = "F2")) +
facet_wrap(~labels + sex)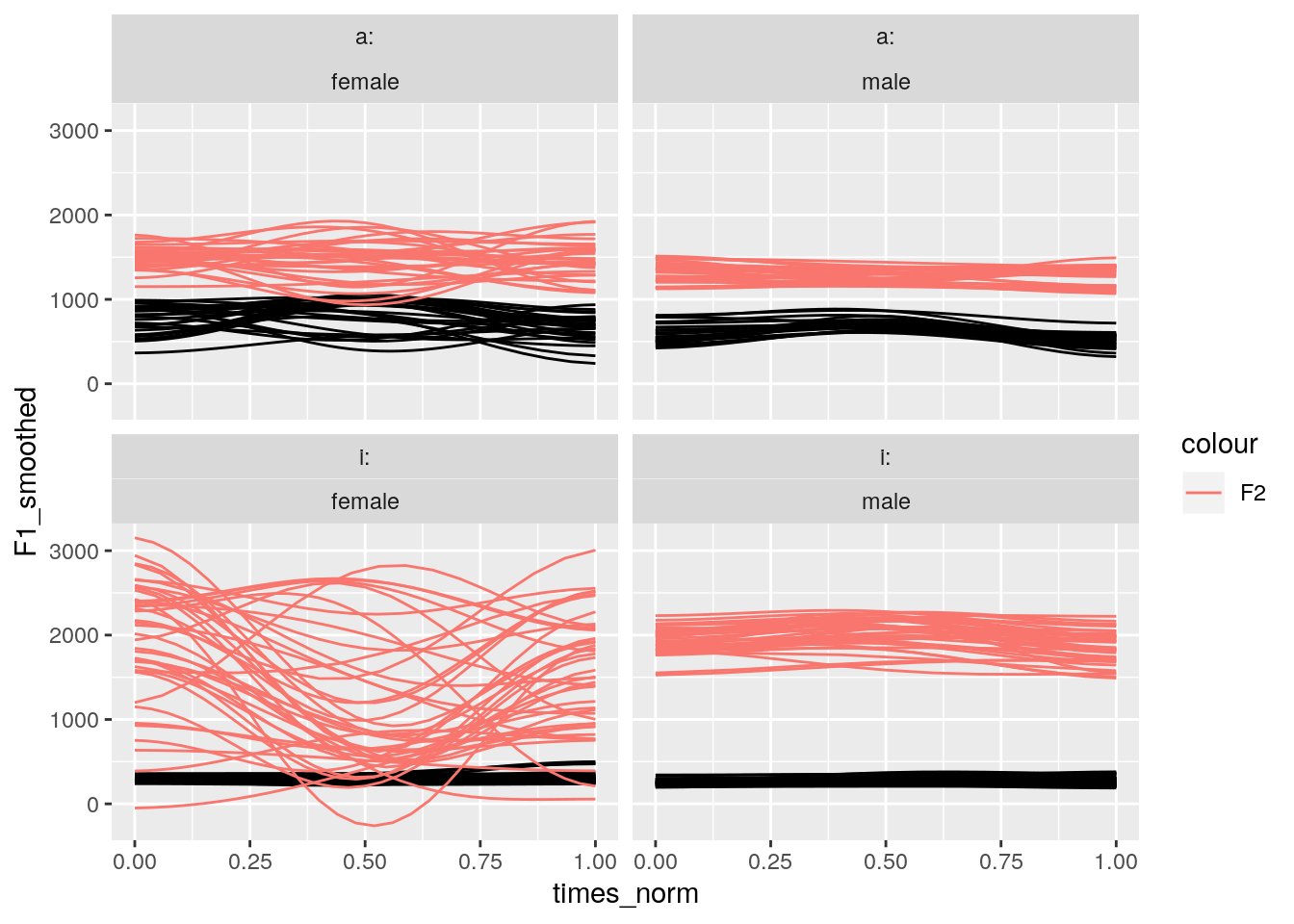
Fricative spectral analysis
- Q: How do we add a SSFF track to the emuDB called ‘dft’ and also calculate the SSFF files using the wrassp function dftSpectrum?
- A:
# ignore error if track already exists
add_ssffTrackDefinition(kiel_db,
name = "dft",
onTheFlyFunctionName = "dftSpectrum")## Error in add_ssffTrackDefinition(kiel_db, name = "dft", onTheFlyFunctionName = "dftSpectrum"): ssffTrackDefinitions with name dft already exists for emuDB: kielread!- Q: How do we extract the fricatives x|C|s|z|S|f from the database?
- A:
sl = query(kiel_db, "Phonetic == x|C|s|z|S|f")- Q: How do we extract a single spectral slice at the temporal mid-point of the extracted fricatives?
- A:
td = get_trackdata(kiel_db, sl, ssffTrackName = "dft")##
## INFO: parsing 681 dft segments/events
##
|
| | 0%
|
| | 1%
|
|= | 1%
|
|= | 2%
|
|== | 2%
|
|== | 3%
|
|== | 4%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 8%
|
|====== | 9%
|
|======= | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========= | 14%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 16%
|
|============ | 17%
|
|============ | 18%
|
|============= | 18%
|
|============= | 19%
|
|============== | 19%
|
|============== | 20%
|
|============== | 21%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================= | 24%
|
|================= | 25%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 26%
|
|=================== | 27%
|
|=================== | 28%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|=============================== | 45%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 49%
|
|=================================== | 50%
|
|=================================== | 51%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 59%
|
|========================================== | 60%
|
|========================================== | 61%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 69%
|
|================================================= | 70%
|
|================================================= | 71%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 72%
|
|=================================================== | 73%
|
|=================================================== | 74%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|====================================================== | 78%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 82%
|
|========================================================== | 83%
|
|========================================================== | 84%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 92%
|
|================================================================= | 93%
|
|================================================================= | 94%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%
## INFO: adding fs attribute to trackdata$data fieldstd_norm = normalize_length(td)## Warning: Row indexes must be between 0 and the number of rows (11). Use `NA` as row index to obtain a row full of `NA` values.
## This warning is displayed once per session.td_norm_mid = td_norm %>% filter(times_norm == 0.5)- Q: How do we visualize a averaged slice per fricative type into a single panel?
- A:
td_norm_mid_long = convert_wideToLong(td_norm_mid, calcFreqs = T)
td_norm_mid_long_mean = td_norm_mid_long %>%
group_by(labels, freq) %>%
summarise(track_value = mean(track_value))
ggplot(td_norm_mid_long_mean) +
aes(x = freq, y = track_value, col = labels) +
geom_line()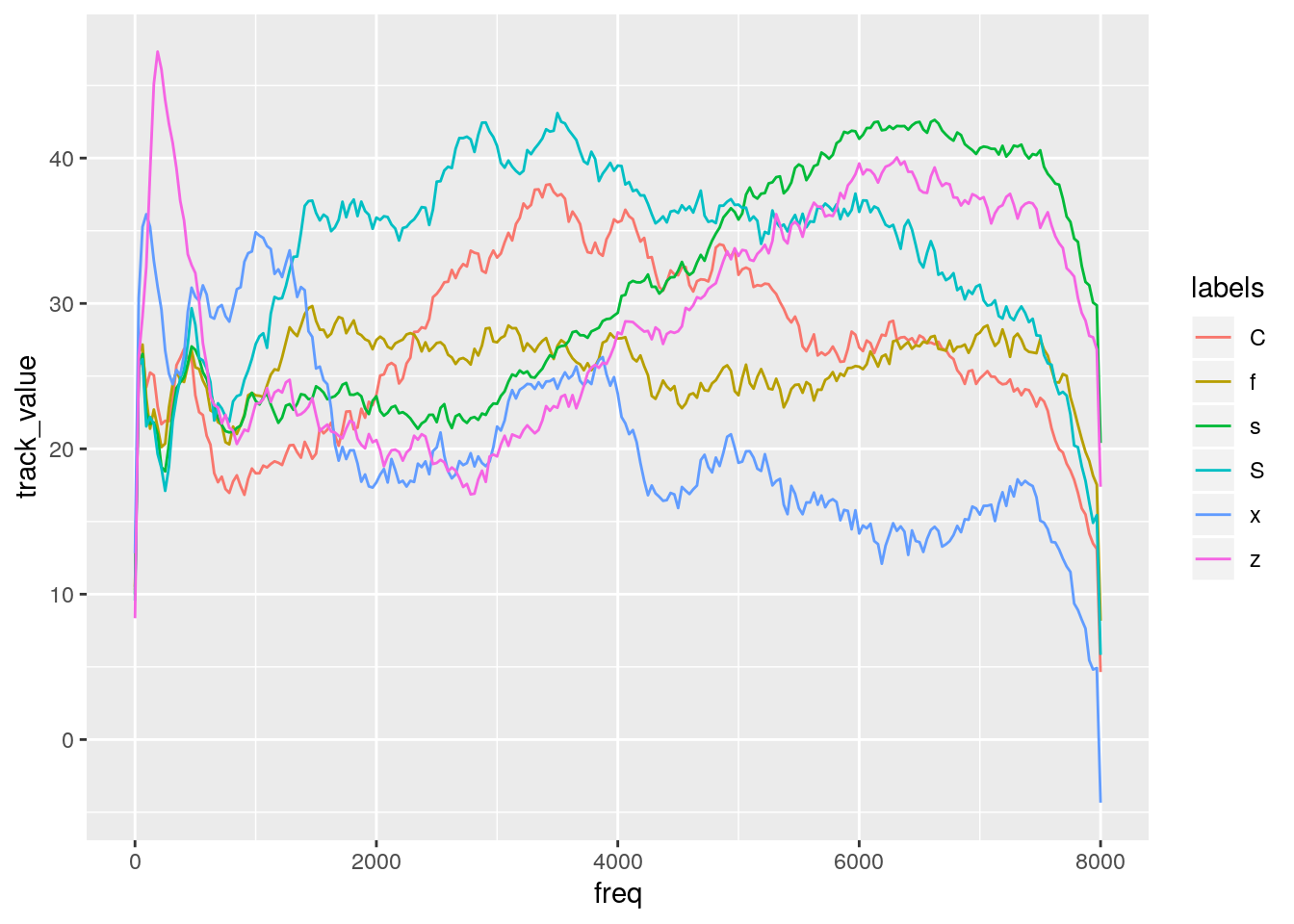
- Q: How do we visualize the dct-smoothed (m=4) spectral slices (one panel per fricative)?
- A:
td_norm_mid_long_smoothed = td_norm_mid_long %>%
group_by(labels) %>%
mutate(dct_smoothed = emuR::dct(track_value, m = 4, fit = T))
ggplot(td_norm_mid_long_smoothed) + # this takes 4 ever to render which is why eval is set to FALSE
aes(x = freq, y = dct_smoothed, col = labels) +
geom_line() +
facet_wrap( ~ sl_rowIdx + labels)Spectral moments analysis
Omitted due to time constraints